Applications
BMSCS - Bayesian Model Selection under Constraints
GitHub Repository Documentation Start App Start App (Beta)
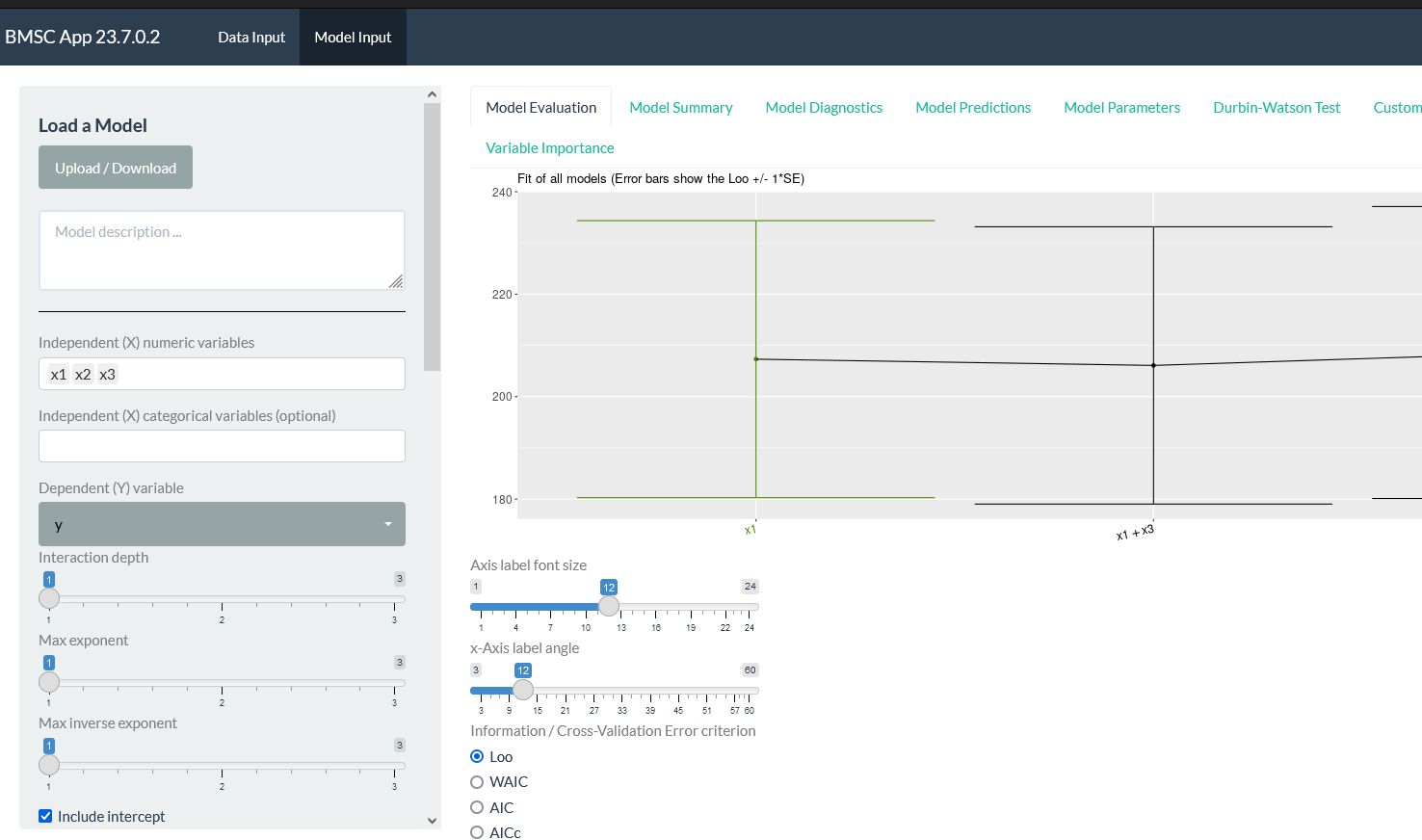
BMSC is a Bayesian model selection algorithm used for causal analysis and other applications.
1. Install the software Docker
Download installation files from one of the links below and follow installation instructions:
After Docker is installed you can pull & run the app manually or use the all-in-one-app.sh-Script.
2. Download and install docker image of the app
This image contains all elements necessary for you to run the app from a web browser. Run this code in a local terminal
Open a terminal (command line):
- Windows command line:
- Open the Start menu or press the
Windows key+R; - Type cmd or cmd.exe in the Run command box;
- Press Enter.
- Open the Start menu or press the
- MacOS: open the Terminal app.
- Linux: most Linux systems use the same default keyboard shortcut to start the command line:
Ctrl-Alt-TorSuper-T
Copy paste the text below into the terminal and press Enter:
docker pull ghcr.io/pandora-isomemo/bmscs:main3. Run the application in Docker
Steps 1 and 2 install the app. To run the app at any time after the installation open a terminal (as described in point 2) copy paste the text below into the terminal and press Enter. Wait for a notification that the app is in “listening” mode.
docker run -p 3838:3838 ghcr.io/pandora-isomemo/bmscs:mainIf the app is shutdown on Docker or if the terminal is closed the app will no longer work in your web browser (see point 4).
4. Display the app in a web browser
Once the app is running in Docker you need to display it in a web browser. For this, copy-paste the address below into your web browser’s address input and press Enter.
http://127.0.0.1:3838/Troubleshooting
- You may not be able to install Docker due a lack of administrative rights. If so, contact your IT department to grant installation rights.
- While trying to use Docker on Windows you may receive the message that the “Windows Subsystem for Linux” is missing. To install it follow these instructions
- While trying to use Docker on Windows you may receive the error “Hardware assisted virtualization and data execution protection must be enabled in the BIOS”. This requires you to access your computer BIOS to enable “hardware virtualization”. Mode of access and BIOS menu options vary with computer model.
- In some problematic cases, you may consider using an older version of Docker. See: https://docs.docker.com/desktop/release-notes/
Install R
Add Repositories
Open a local R Console and add these two additional Repositories.
options(
repos = c(
getOption("repos"),
PANDORA = "https://Pandora-IsoMemo.github.io/drat/"
INWTlab = "https://inwtlab.github.io/drat/"
)
)Install Package
Load and Install the R-Package
install.packages("BMSCS")Start the app
Load and attach package
library("BMSCS")Run the app
BMSCS::startApplication()BNR
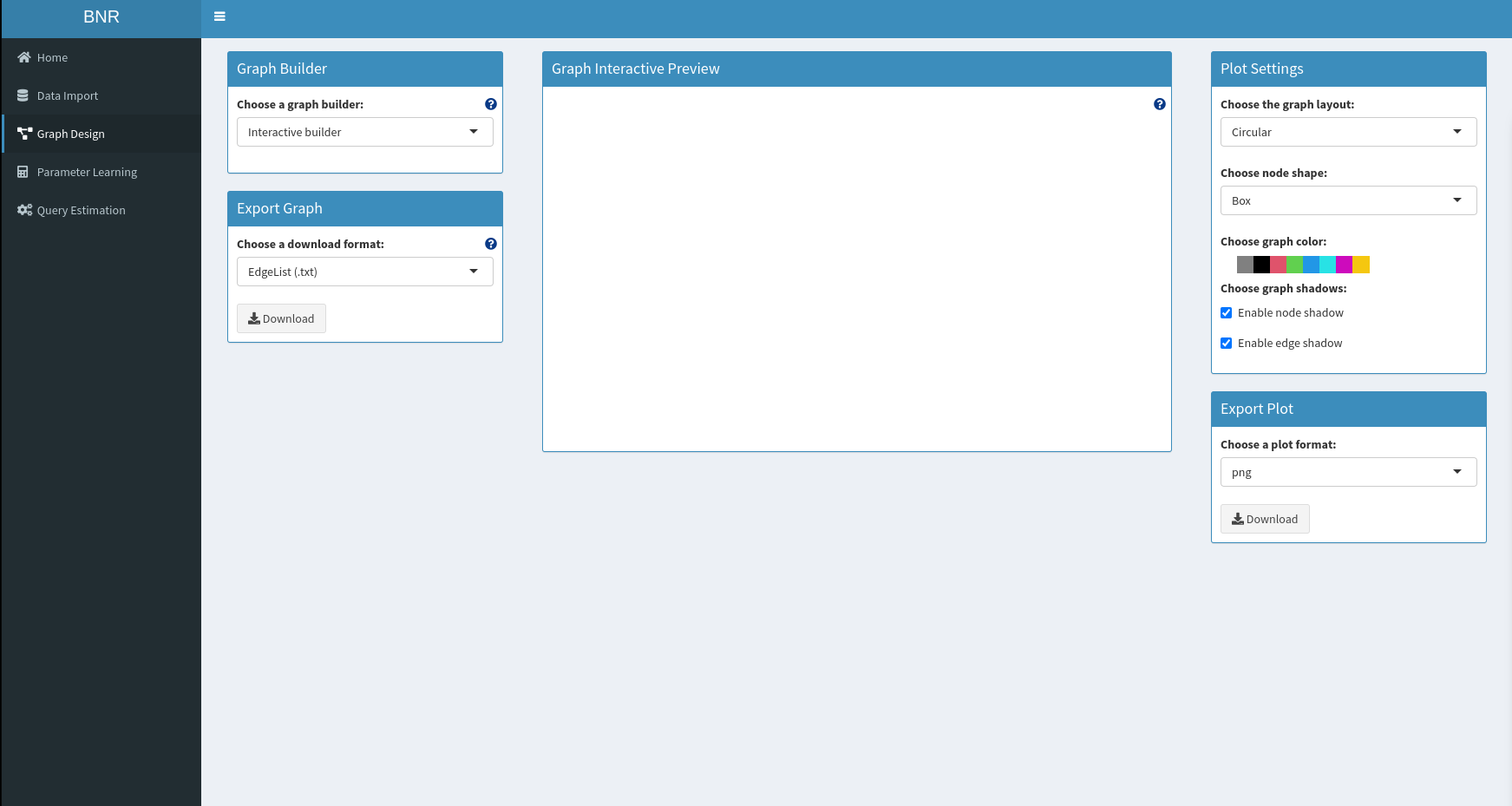
This project leverages graphical models to perform queries with constrained bayesian estimation. This work is developed as a Shiny app in order to provide an easy and clean interaction.
1. Install the software Docker
Download installation files from one of the links below and follow installation instructions:
After Docker is installed you can pull & run the app manually or use the all-in-one-app.sh-Script.
2. Download and install docker image of the app
This image contains all elements necessary for you to run the app from a web browser. Run this code in a local terminal
Open a terminal (command line):
- Windows command line:
- Open the Start menu or press the
Windows key+R; - Type cmd or cmd.exe in the Run command box;
- Press Enter.
- Open the Start menu or press the
- MacOS: open the Terminal app.
- Linux: most Linux systems use the same default keyboard shortcut to start the command line:
Ctrl-Alt-TorSuper-T
Copy paste the text below into the terminal and press Enter:
docker pull ghcr.io/pandora-isomemo/bnr:main3. Run the application in Docker
Steps 1 and 2 install the app. To run the app at any time after the installation open a terminal (as described in point 2) copy paste the text below into the terminal and press Enter. Wait for a notification that the app is in “listening” mode.
docker run -p 3838:3838 ghcr.io/pandora-isomemo/bnr:mainIf the app is shutdown on Docker or if the terminal is closed the app will no longer work in your web browser (see point 4).
4. Display the app in a web browser
Once the app is running in Docker you need to display it in a web browser. For this, copy-paste the address below into your web browser’s address input and press Enter.
http://127.0.0.1:3838/Troubleshooting
- You may not be able to install Docker due a lack of administrative rights. If so, contact your IT department to grant installation rights.
- While trying to use Docker on Windows you may receive the message that the “Windows Subsystem for Linux” is missing. To install it follow these instructions
- While trying to use Docker on Windows you may receive the error “Hardware assisted virtualization and data execution protection must be enabled in the BIOS”. This requires you to access your computer BIOS to enable “hardware virtualization”. Mode of access and BIOS menu options vary with computer model.
- In some problematic cases, you may consider using an older version of Docker. See: https://docs.docker.com/desktop/release-notes/
Install R
Add Repositories
Open a local R Console and add these two additional Repositories.
options(
repos = c(
getOption("repos"),
PANDORA = "https://Pandora-IsoMemo.github.io/drat/"
INWTlab = "https://inwtlab.github.io/drat/"
)
)Use Renv
Install R dependencies with renv
# install.packages("renv")
renv::restore()Run the shinyapp
Run the application by executing:
shiny::runApp()Bpred - Bayesian Prediction
GitHub Repository Documentation Start App Start App (Beta)
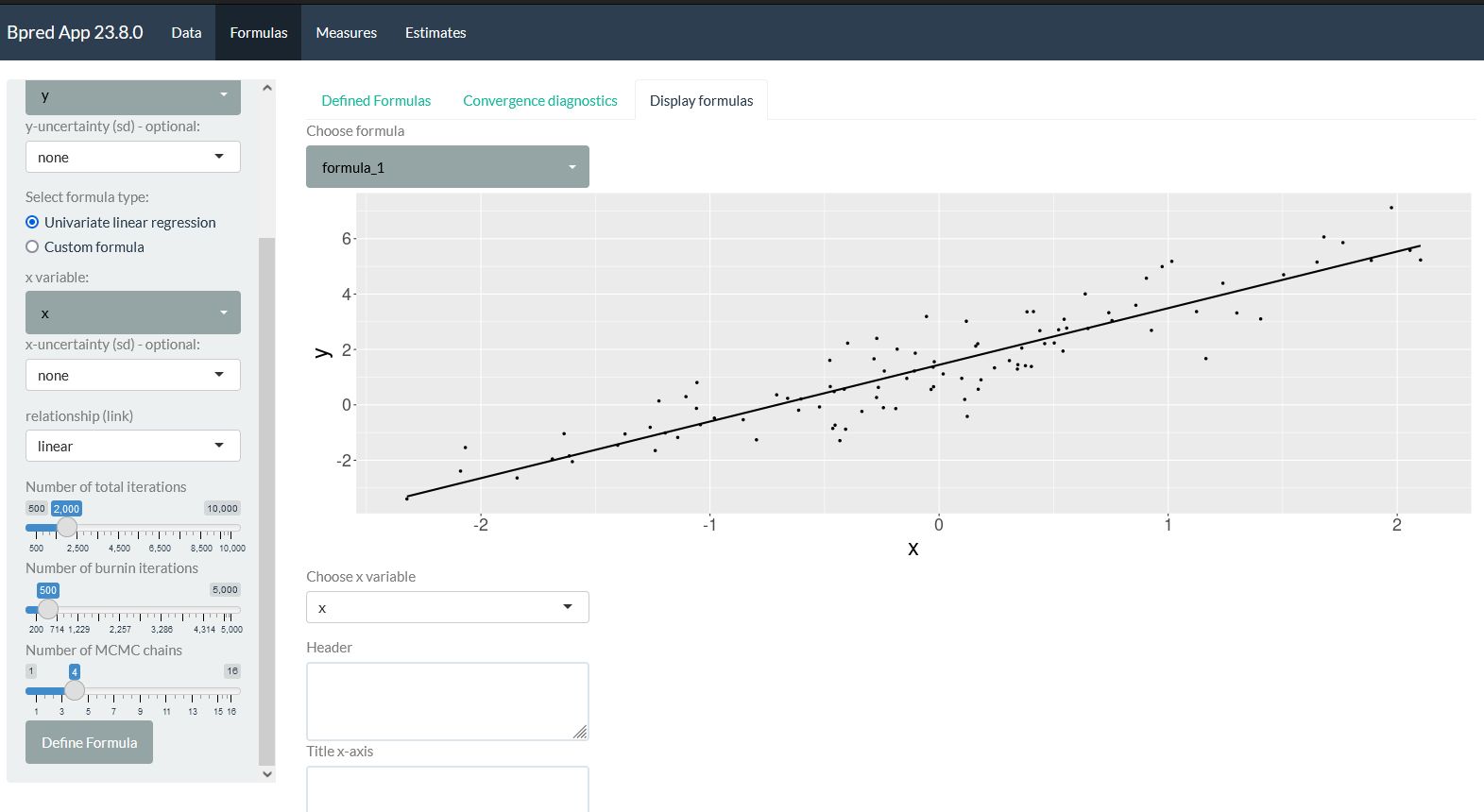
Bayesian multivariate regression application.
1. Install the software Docker
Download installation files from one of the links below and follow installation instructions:
After Docker is installed you can pull & run the app manually or use the all-in-one-app.sh-Script.
2. Download and install docker image of the app
This image contains all elements necessary for you to run the app from a web browser. Run this code in a local terminal
Open a terminal (command line):
- Windows command line:
- Open the Start menu or press the
Windows key+R; - Type cmd or cmd.exe in the Run command box;
- Press Enter.
- Open the Start menu or press the
- MacOS: open the Terminal app.
- Linux: most Linux systems use the same default keyboard shortcut to start the command line:
Ctrl-Alt-TorSuper-T
Copy paste the text below into the terminal and press Enter:
docker pull ghcr.io/pandora-isomemo/bpred:main3. Run the application in Docker
Steps 1 and 2 install the app. To run the app at any time after the installation open a terminal (as described in point 2) copy paste the text below into the terminal and press Enter. Wait for a notification that the app is in “listening” mode.
docker run -p 3838:3838 ghcr.io/pandora-isomemo/bpred:mainIf the app is shutdown on Docker or if the terminal is closed the app will no longer work in your web browser (see point 4).
4. Display the app in a web browser
Once the app is running in Docker you need to display it in a web browser. For this, copy-paste the address below into your web browser’s address input and press Enter.
http://127.0.0.1:3838/Troubleshooting
- You may not be able to install Docker due a lack of administrative rights. If so, contact your IT department to grant installation rights.
- While trying to use Docker on Windows you may receive the message that the “Windows Subsystem for Linux” is missing. To install it follow these instructions
- While trying to use Docker on Windows you may receive the error “Hardware assisted virtualization and data execution protection must be enabled in the BIOS”. This requires you to access your computer BIOS to enable “hardware virtualization”. Mode of access and BIOS menu options vary with computer model.
- In some problematic cases, you may consider using an older version of Docker. See: https://docs.docker.com/desktop/release-notes/
Install R
Add Repositories
Open a local R Console and add these two additional Repositories.
options(
repos = c(
getOption("repos"),
PANDORA = "https://Pandora-IsoMemo.github.io/drat/"
INWTlab = "https://inwtlab.github.io/drat/"
)
)Install Package
Load and Install the R-Package
install.packages("Bpred")Start the app
Load and attach package
library("Bpred")Run the app
Bpred::startApplication()CausalR
GitHub Repository Video Tutorial Start App Start App (Beta)
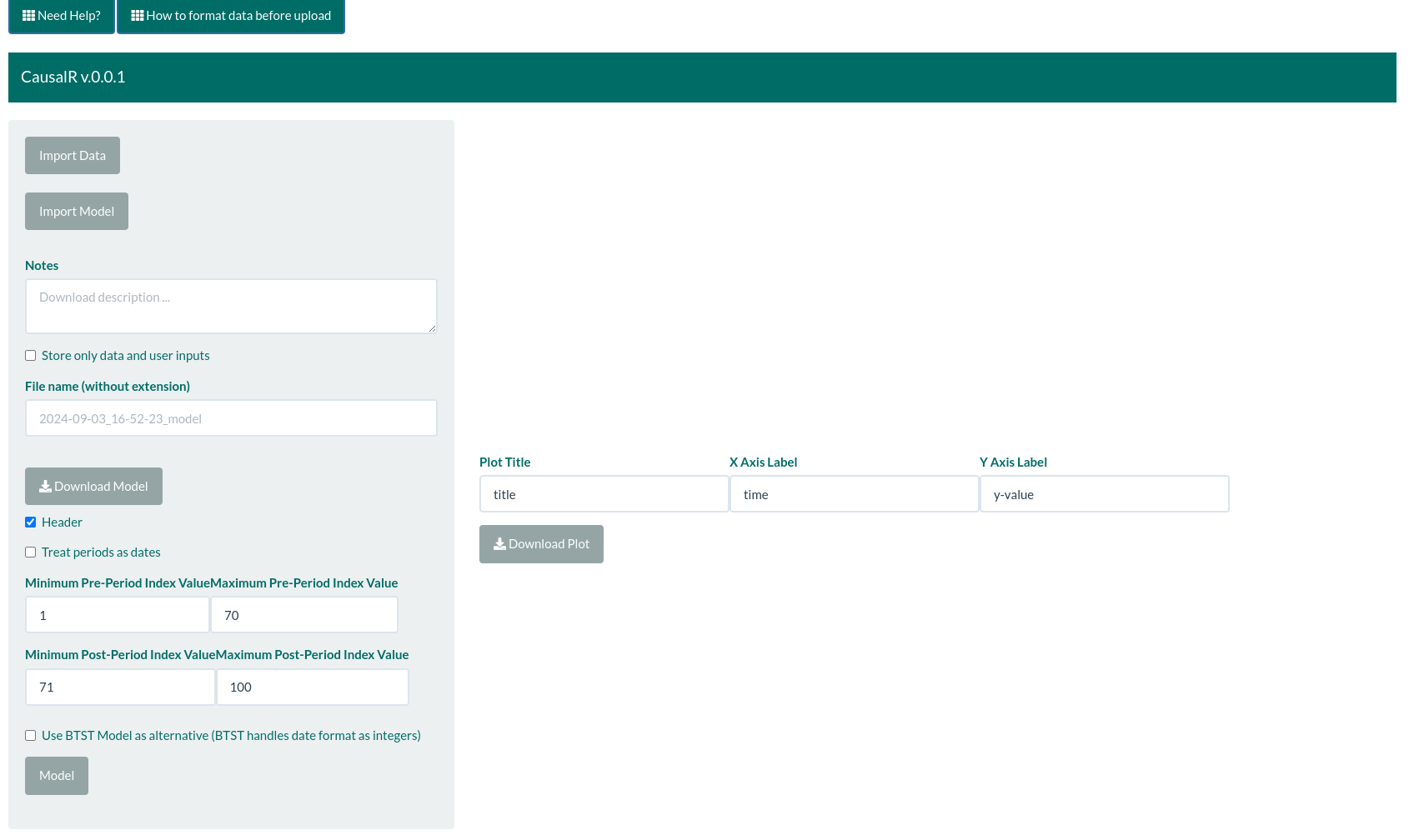
CausalR is an R-Shiny user-friendly interface for CausalImpact R package, which leverages the Bayesian modeling capabilities to estimate the causal effect of an intervention on a time series.
1. Install the software Docker
Download installation files from one of the links below and follow installation instructions:
After Docker is installed you can pull & run the app manually or use the all-in-one-app.sh-Script.
2. Download and install docker image of the app
This image contains all elements necessary for you to run the app from a web browser. Run this code in a local terminal
Open a terminal (command line):
- Windows command line:
- Open the Start menu or press the
Windows key+R; - Type cmd or cmd.exe in the Run command box;
- Press Enter.
- Open the Start menu or press the
- MacOS: open the Terminal app.
- Linux: most Linux systems use the same default keyboard shortcut to start the command line:
Ctrl-Alt-TorSuper-T
Copy paste the text below into the terminal and press Enter:
docker pull ghcr.io/pandora-isomemo/causalr:main3. Run the application in Docker
Steps 1 and 2 install the app. To run the app at any time after the installation open a terminal (as described in point 2) copy paste the text below into the terminal and press Enter. Wait for a notification that the app is in “listening” mode.
docker run -p 3838:3838 ghcr.io/pandora-isomemo/causalr:mainIf the app is shutdown on Docker or if the terminal is closed the app will no longer work in your web browser (see point 4).
4. Display the app in a web browser
Once the app is running in Docker you need to display it in a web browser. For this, copy-paste the address below into your web browser’s address input and press Enter.
http://127.0.0.1:3838/Troubleshooting
- You may not be able to install Docker due a lack of administrative rights. If so, contact your IT department to grant installation rights.
- While trying to use Docker on Windows you may receive the message that the “Windows Subsystem for Linux” is missing. To install it follow these instructions
- While trying to use Docker on Windows you may receive the error “Hardware assisted virtualization and data execution protection must be enabled in the BIOS”. This requires you to access your computer BIOS to enable “hardware virtualization”. Mode of access and BIOS menu options vary with computer model.
- In some problematic cases, you may consider using an older version of Docker. See: https://docs.docker.com/desktop/release-notes/
Install R
Add Repositories
Open a local R Console and add these two additional Repositories.
options(
repos = c(
getOption("repos"),
PANDORA = "https://Pandora-IsoMemo.github.io/drat/"
INWTlab = "https://inwtlab.github.io/drat/"
)
)Install Package
Load and Install the R-Package
install.packages("CausalR")Start the app
Load and attach package
library("CausalR")Run the app
CausalR::startApplication()ChangeR
GitHub Repository Documentation Start App Start App (Beta)
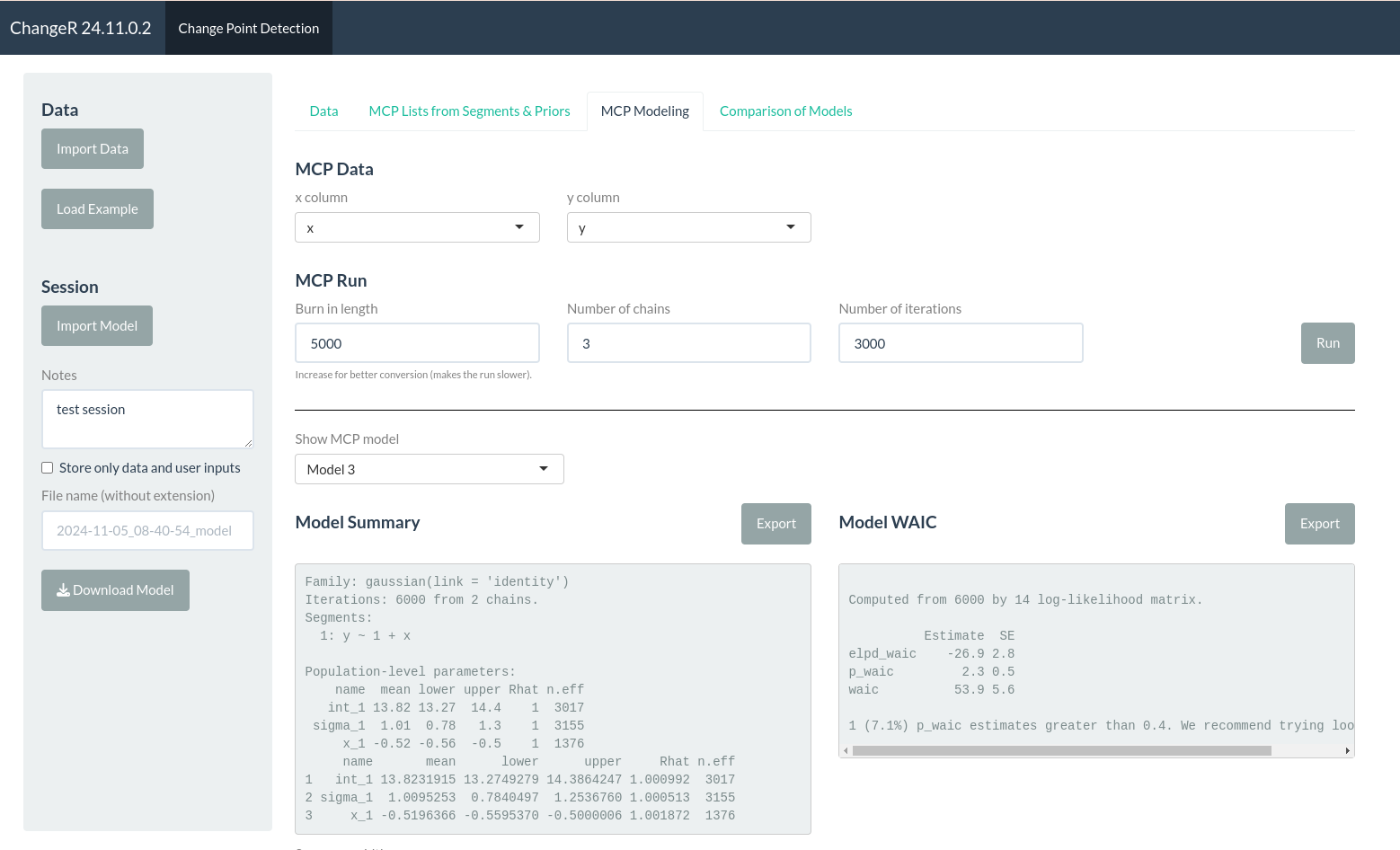
An App to display and identify isotopic change points in human tooth sections.
1. Install the software Docker
Download installation files from one of the links below and follow installation instructions:
After Docker is installed you can pull & run the app manually or use the all-in-one-app.sh-Script.
2. Download and install docker image of the app
This image contains all elements necessary for you to run the app from a web browser. Run this code in a local terminal
Open a terminal (command line):
- Windows command line:
- Open the Start menu or press the
Windows key+R; - Type cmd or cmd.exe in the Run command box;
- Press Enter.
- Open the Start menu or press the
- MacOS: open the Terminal app.
- Linux: most Linux systems use the same default keyboard shortcut to start the command line:
Ctrl-Alt-TorSuper-T
Copy paste the text below into the terminal and press Enter:
docker pull ghcr.io/pandora-isomemo/changer:main3. Run the application in Docker
Steps 1 and 2 install the app. To run the app at any time after the installation open a terminal (as described in point 2) copy paste the text below into the terminal and press Enter. Wait for a notification that the app is in “listening” mode.
docker run -p 3838:3838 ghcr.io/pandora-isomemo/changer:mainIf the app is shutdown on Docker or if the terminal is closed the app will no longer work in your web browser (see point 4).
4. Display the app in a web browser
Once the app is running in Docker you need to display it in a web browser. For this, copy-paste the address below into your web browser’s address input and press Enter.
http://127.0.0.1:3838/Troubleshooting
- You may not be able to install Docker due a lack of administrative rights. If so, contact your IT department to grant installation rights.
- While trying to use Docker on Windows you may receive the message that the “Windows Subsystem for Linux” is missing. To install it follow these instructions
- While trying to use Docker on Windows you may receive the error “Hardware assisted virtualization and data execution protection must be enabled in the BIOS”. This requires you to access your computer BIOS to enable “hardware virtualization”. Mode of access and BIOS menu options vary with computer model.
- In some problematic cases, you may consider using an older version of Docker. See: https://docs.docker.com/desktop/release-notes/
ChangeR utilizes JAGS MCMC library. This has been tested only on Linux (Ubuntu). For Windows & Mac user, you can either use the dockerized Version of the App or refer to the offical installation manual of JAGS.
Install R
Add Repositories
Open a local R Console and add these two additional Repositories.
options(
repos = c(
getOption("repos"),
PANDORA = "https://Pandora-IsoMemo.github.io/drat/"
INWTlab = "https://inwtlab.github.io/drat/"
)
)Install Package
Load and Install the R-Package
install.packages("ChangeR")Start the app
Load and attach package
library("ChangeR")Run the app
ChangeR::startApplication()DSSM - Data Search and Spatiotemporal Modelling
GitHub Repository Documentation Video Tutorial Start App Start App (Beta)
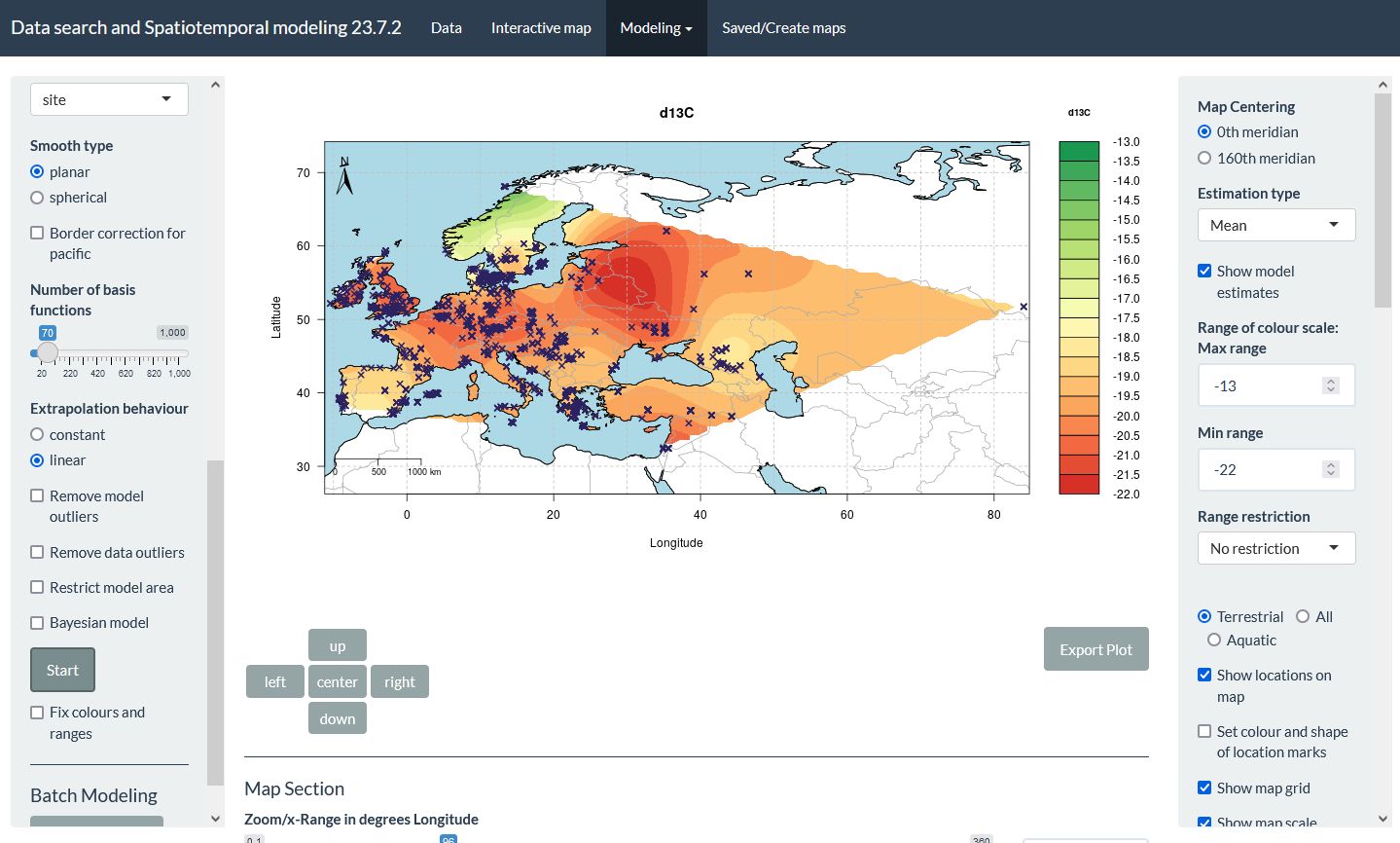
In addition to allowing users to query Pandora data includes different types of Bayesian and non-Bayesian spatiotemporal models.
1. Install the software Docker
Download installation files from one of the links below and follow installation instructions:
After Docker is installed you can pull & run the app manually or use the all-in-one-app.sh-Script.
2. Download and install docker image of the app
This image contains all elements necessary for you to run the app from a web browser. Run this code in a local terminal
Open a terminal (command line):
- Windows command line:
- Open the Start menu or press the
Windows key+R; - Type cmd or cmd.exe in the Run command box;
- Press Enter.
- Open the Start menu or press the
- MacOS: open the Terminal app.
- Linux: most Linux systems use the same default keyboard shortcut to start the command line:
Ctrl-Alt-TorSuper-T
Copy paste the text below into the terminal and press Enter:
docker pull ghcr.io/pandora-isomemo/dssm:main3. Run the application in Docker
Steps 1 and 2 install the app. To run the app at any time after the installation open a terminal (as described in point 2) copy paste the text below into the terminal and press Enter. Wait for a notification that the app is in “listening” mode.
docker run -p 3838:3838 ghcr.io/pandora-isomemo/dssm:mainIf the app is shutdown on Docker or if the terminal is closed the app will no longer work in your web browser (see point 4).
4. Display the app in a web browser
Once the app is running in Docker you need to display it in a web browser. For this, copy-paste the address below into your web browser’s address input and press Enter.
http://127.0.0.1:3838/Troubleshooting
- You may not be able to install Docker due a lack of administrative rights. If so, contact your IT department to grant installation rights.
- While trying to use Docker on Windows you may receive the message that the “Windows Subsystem for Linux” is missing. To install it follow these instructions
- While trying to use Docker on Windows you may receive the error “Hardware assisted virtualization and data execution protection must be enabled in the BIOS”. This requires you to access your computer BIOS to enable “hardware virtualization”. Mode of access and BIOS menu options vary with computer model.
- In some problematic cases, you may consider using an older version of Docker. See: https://docs.docker.com/desktop/release-notes/
Install R
Add Repositories
Open a local R Console and add these two additional Repositories.
options(
repos = c(
getOption("repos"),
PANDORA = "https://Pandora-IsoMemo.github.io/drat/"
INWTlab = "https://inwtlab.github.io/drat/"
)
)Install Package
Load and Install the R-Package
install.packages("DSSM")Start the app
Load and attach package
library("DSSM")Run the app
DSSM::startApplication()MapR
GitHub Repository Documentation Start App Start App (Beta)
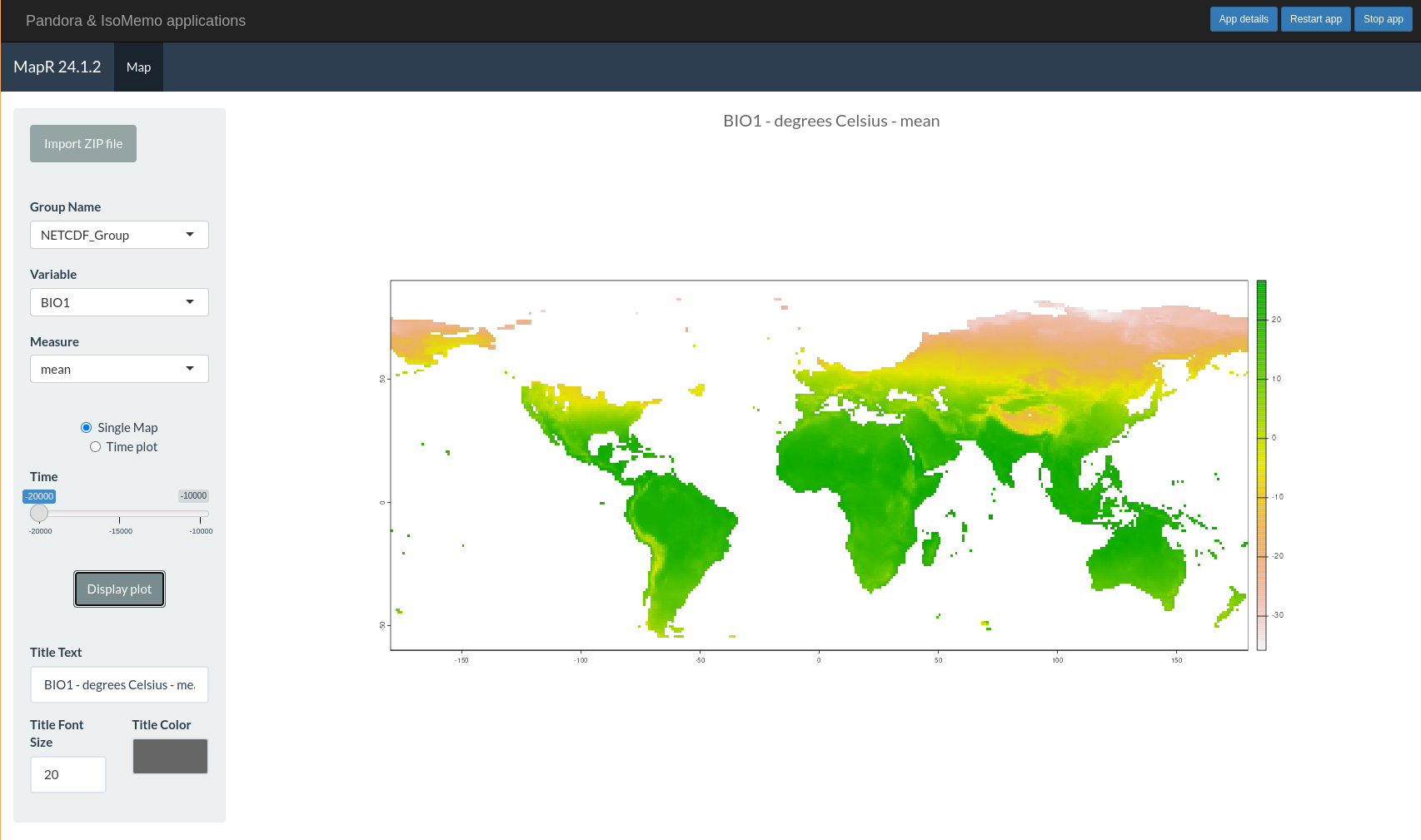
MapR displays temporal and temperature graphical files for Isomemo.
1. Install the software Docker
Download installation files from one of the links below and follow installation instructions:
After Docker is installed you can pull & run the app manually or use the all-in-one-app.sh-Script.
2. Download and install docker image of the app
This image contains all elements necessary for you to run the app from a web browser. Run this code in a local terminal
Open a terminal (command line):
- Windows command line:
- Open the Start menu or press the
Windows key+R; - Type cmd or cmd.exe in the Run command box;
- Press Enter.
- Open the Start menu or press the
- MacOS: open the Terminal app.
- Linux: most Linux systems use the same default keyboard shortcut to start the command line:
Ctrl-Alt-TorSuper-T
Copy paste the text below into the terminal and press Enter:
docker pull ghcr.io/pandora-isomemo/mapr:main3. Run the application in Docker
Steps 1 and 2 install the app. To run the app at any time after the installation open a terminal (as described in point 2) copy paste the text below into the terminal and press Enter. Wait for a notification that the app is in “listening” mode.
docker run -p 3838:3838 ghcr.io/pandora-isomemo/mapr:mainIf the app is shutdown on Docker or if the terminal is closed the app will no longer work in your web browser (see point 4).
4. Display the app in a web browser
Once the app is running in Docker you need to display it in a web browser. For this, copy-paste the address below into your web browser’s address input and press Enter.
http://127.0.0.1:3838/Troubleshooting
- You may not be able to install Docker due a lack of administrative rights. If so, contact your IT department to grant installation rights.
- While trying to use Docker on Windows you may receive the message that the “Windows Subsystem for Linux” is missing. To install it follow these instructions
- While trying to use Docker on Windows you may receive the error “Hardware assisted virtualization and data execution protection must be enabled in the BIOS”. This requires you to access your computer BIOS to enable “hardware virtualization”. Mode of access and BIOS menu options vary with computer model.
- In some problematic cases, you may consider using an older version of Docker. See: https://docs.docker.com/desktop/release-notes/
Install R
Add Repositories
Open a local R Console and add these two additional Repositories.
options(
repos = c(
getOption("repos"),
PANDORA = "https://Pandora-IsoMemo.github.io/drat/"
INWTlab = "https://inwtlab.github.io/drat/"
)
)Install Package
Load and Install the R-Package
install.packages("MapR")Start the app
Load and attach package
library("MapR")Run the app
MapR::startApplication()OsteoBioR
GitHub Repository Documentation Start App Start App (Beta)
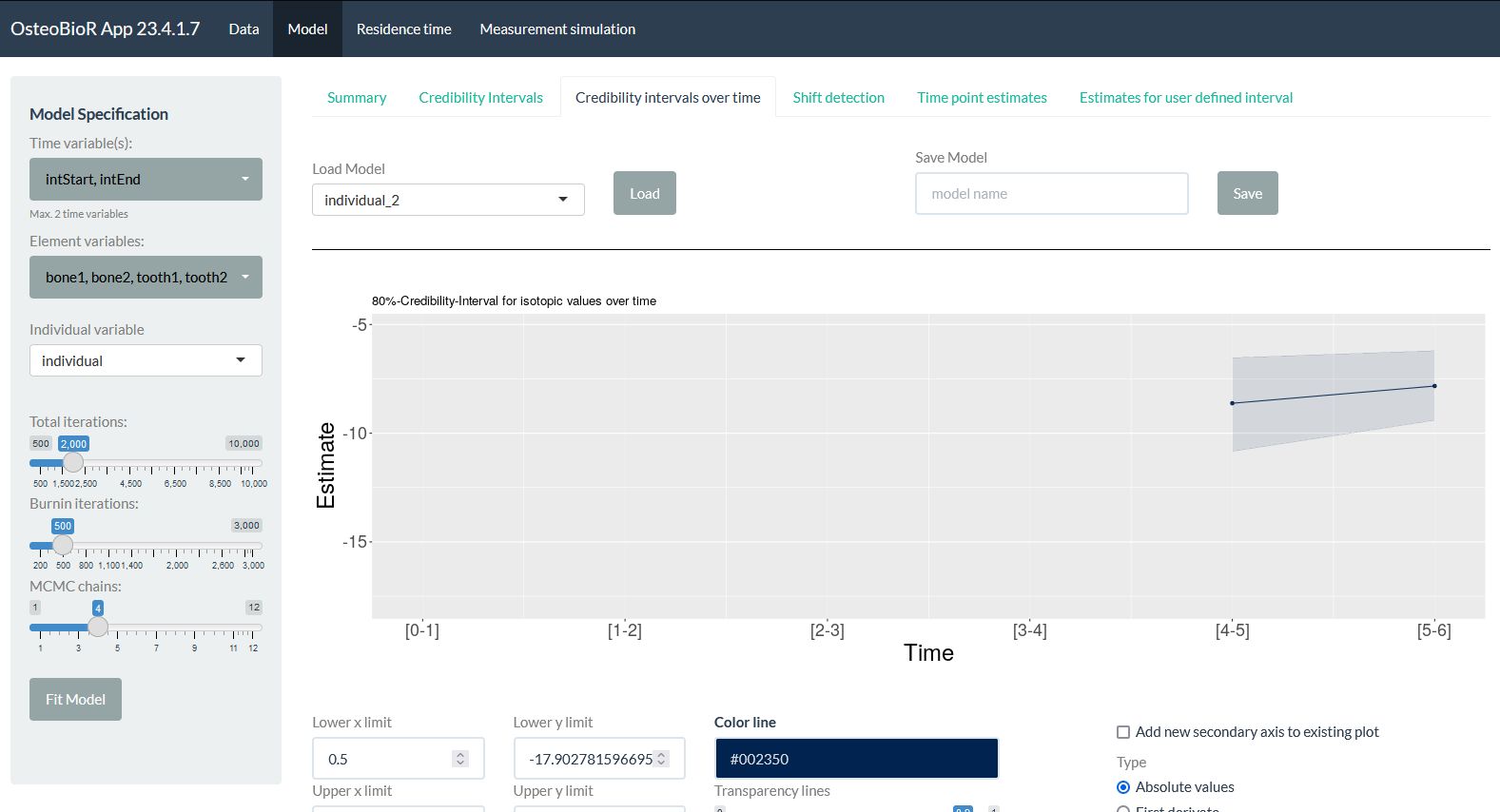
OsteoBioR is used to model sequential data where each measurement is known to cover a time interval (integration time). This can be used, among others, to model isotopic temporal trajectories from isotopic measurements taken in multiple living tissues.
1. Install the software Docker
Download installation files from one of the links below and follow installation instructions:
After Docker is installed you can pull & run the app manually or use the all-in-one-app.sh-Script.
2. Download and install docker image of the app
This image contains all elements necessary for you to run the app from a web browser. Run this code in a local terminal
Open a terminal (command line):
- Windows command line:
- Open the Start menu or press the
Windows key+R; - Type cmd or cmd.exe in the Run command box;
- Press Enter.
- Open the Start menu or press the
- MacOS: open the Terminal app.
- Linux: most Linux systems use the same default keyboard shortcut to start the command line:
Ctrl-Alt-TorSuper-T
Copy paste the text below into the terminal and press Enter:
docker pull ghcr.io/pandora-isomemo/osteobior:main3. Run the application in Docker
Steps 1 and 2 install the app. To run the app at any time after the installation open a terminal (as described in point 2) copy paste the text below into the terminal and press Enter. Wait for a notification that the app is in “listening” mode.
docker run -p 3838:3838 ghcr.io/pandora-isomemo/osteobior:mainIf the app is shutdown on Docker or if the terminal is closed the app will no longer work in your web browser (see point 4).
4. Display the app in a web browser
Once the app is running in Docker you need to display it in a web browser. For this, copy-paste the address below into your web browser’s address input and press Enter.
http://127.0.0.1:3838/Troubleshooting
- You may not be able to install Docker due a lack of administrative rights. If so, contact your IT department to grant installation rights.
- While trying to use Docker on Windows you may receive the message that the “Windows Subsystem for Linux” is missing. To install it follow these instructions
- While trying to use Docker on Windows you may receive the error “Hardware assisted virtualization and data execution protection must be enabled in the BIOS”. This requires you to access your computer BIOS to enable “hardware virtualization”. Mode of access and BIOS menu options vary with computer model.
- In some problematic cases, you may consider using an older version of Docker. See: https://docs.docker.com/desktop/release-notes/
Install R
Add Repositories
Open a local R Console and add these two additional Repositories.
options(
repos = c(
getOption("repos"),
PANDORA = "https://Pandora-IsoMemo.github.io/drat/"
INWTlab = "https://inwtlab.github.io/drat/"
)
)Generate C++ Code
(Re-)generate R/rstanmodel.R and the C++ Source Code:
# install.packages("rstantools")
rstantools::rstan_config()Install Package
Load and Install the R-Package
install.packages("OsteoBioR")Start the app
Load and attach package
library("OsteoBioR")Run the app
OsteoBioR::startApplication()PlotR
GitHub Repository Documentation Start App Start App (Beta)
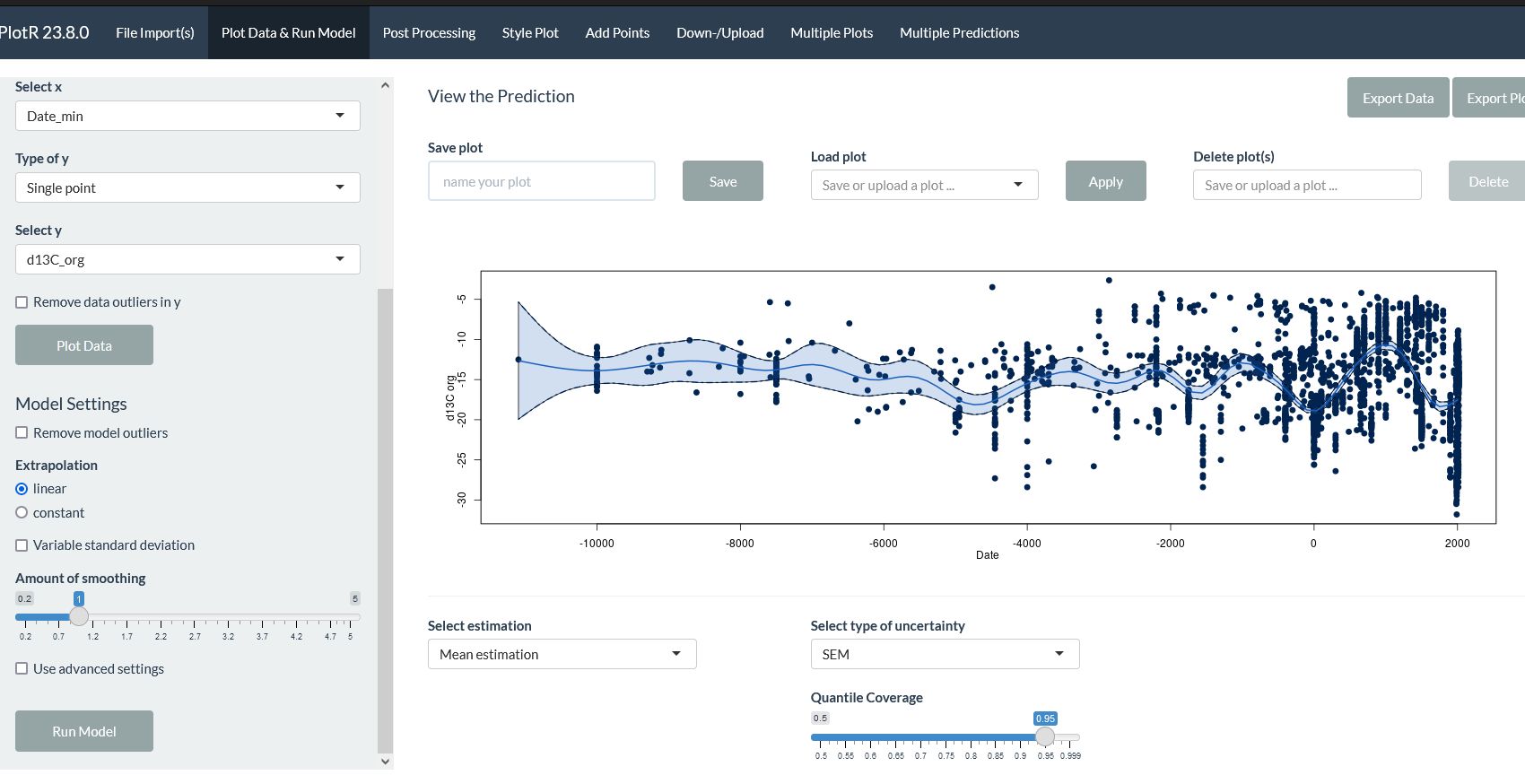
PlotR produces Bayesian smoothed plots.
1. Install the software Docker
Download installation files from one of the links below and follow installation instructions:
After Docker is installed you can pull & run the app manually or use the all-in-one-app.sh-Script.
2. Download and install docker image of the app
This image contains all elements necessary for you to run the app from a web browser. Run this code in a local terminal
Open a terminal (command line):
- Windows command line:
- Open the Start menu or press the
Windows key+R; - Type cmd or cmd.exe in the Run command box;
- Press Enter.
- Open the Start menu or press the
- MacOS: open the Terminal app.
- Linux: most Linux systems use the same default keyboard shortcut to start the command line:
Ctrl-Alt-TorSuper-T
Copy paste the text below into the terminal and press Enter:
docker pull ghcr.io/pandora-isomemo/plotr:main3. Run the application in Docker
Steps 1 and 2 install the app. To run the app at any time after the installation open a terminal (as described in point 2) copy paste the text below into the terminal and press Enter. Wait for a notification that the app is in “listening” mode.
docker run -p 3838:3838 ghcr.io/pandora-isomemo/plotr:mainIf the app is shutdown on Docker or if the terminal is closed the app will no longer work in your web browser (see point 4).
4. Display the app in a web browser
Once the app is running in Docker you need to display it in a web browser. For this, copy-paste the address below into your web browser’s address input and press Enter.
http://127.0.0.1:3838/Troubleshooting
- You may not be able to install Docker due a lack of administrative rights. If so, contact your IT department to grant installation rights.
- While trying to use Docker on Windows you may receive the message that the “Windows Subsystem for Linux” is missing. To install it follow these instructions
- While trying to use Docker on Windows you may receive the error “Hardware assisted virtualization and data execution protection must be enabled in the BIOS”. This requires you to access your computer BIOS to enable “hardware virtualization”. Mode of access and BIOS menu options vary with computer model.
- In some problematic cases, you may consider using an older version of Docker. See: https://docs.docker.com/desktop/release-notes/
Install R
Add Repositories
Open a local R Console and add these two additional Repositories.
options(
repos = c(
getOption("repos"),
PANDORA = "https://Pandora-IsoMemo.github.io/drat/"
INWTlab = "https://inwtlab.github.io/drat/"
)
)Install Package
Load and Install the R-Package
install.packages("PlotR")Start the app
Load and attach package
library("PlotR")Run the app
PlotR::startApplication()ReSources - Food Reconstruction Using Isotopic Transferred Signals
GitHub Repository Documentation Video Tutorial Start App Start App (Beta)
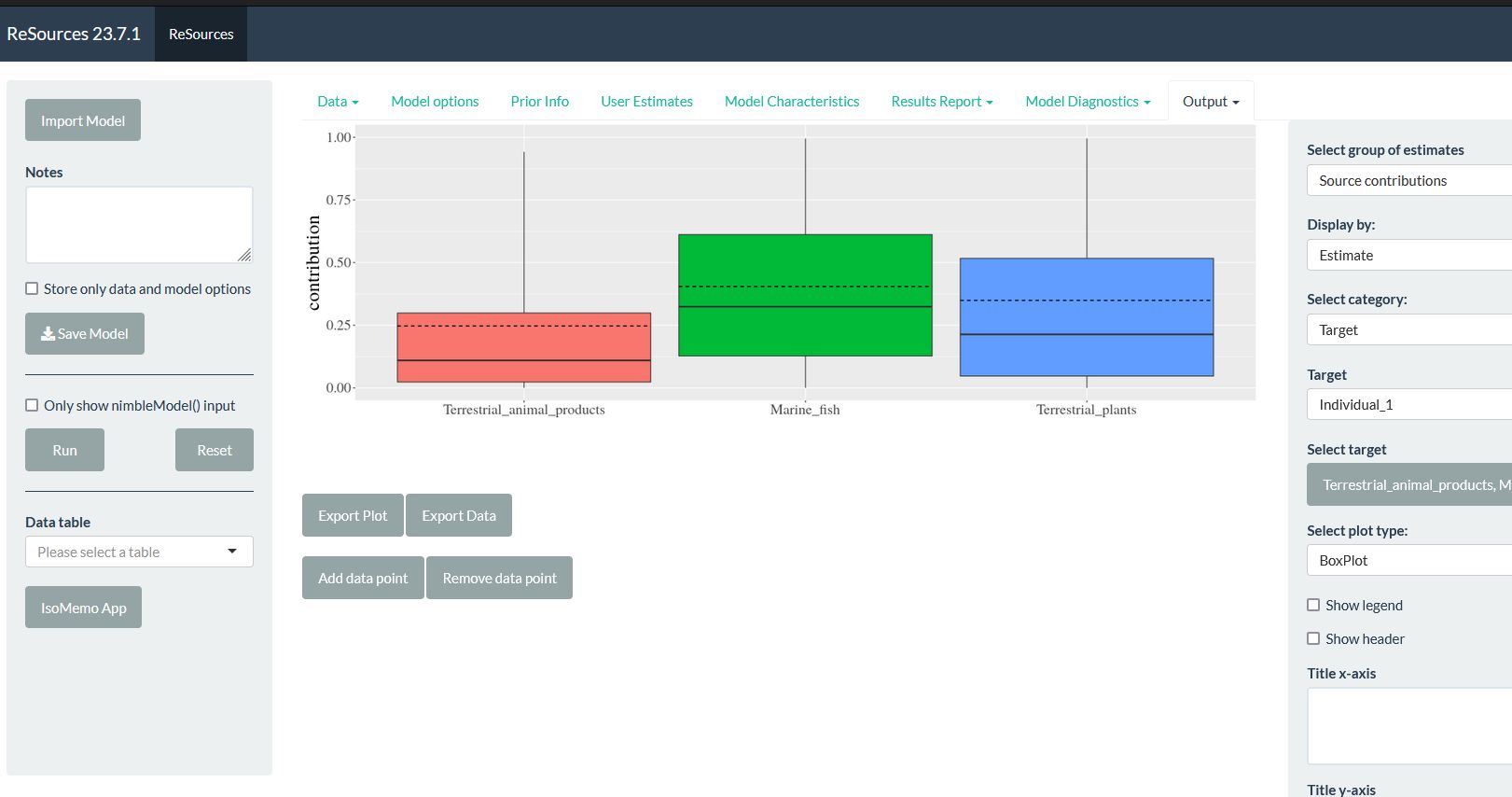
Bayesian mixing model.
1. Install the software Docker
Download installation files from one of the links below and follow installation instructions:
After Docker is installed you can pull & run the app manually or use the all-in-one-app.sh-Script.
2. Download and install docker image of the app
This image contains all elements necessary for you to run the app from a web browser. Run this code in a local terminal
Open a terminal (command line):
- Windows command line:
- Open the Start menu or press the
Windows key+R; - Type cmd or cmd.exe in the Run command box;
- Press Enter.
- Open the Start menu or press the
- MacOS: open the Terminal app.
- Linux: most Linux systems use the same default keyboard shortcut to start the command line:
Ctrl-Alt-TorSuper-T
Copy paste the text below into the terminal and press Enter:
docker pull ghcr.io/pandora-isomemo/resources:main3. Run the application in Docker
Steps 1 and 2 install the app. To run the app at any time after the installation open a terminal (as described in point 2) copy paste the text below into the terminal and press Enter. Wait for a notification that the app is in “listening” mode.
docker run -p 3838:3838 ghcr.io/pandora-isomemo/resources:mainIf the app is shutdown on Docker or if the terminal is closed the app will no longer work in your web browser (see point 4).
4. Display the app in a web browser
Once the app is running in Docker you need to display it in a web browser. For this, copy-paste the address below into your web browser’s address input and press Enter.
http://127.0.0.1:3838/Troubleshooting
- You may not be able to install Docker due a lack of administrative rights. If so, contact your IT department to grant installation rights.
- While trying to use Docker on Windows you may receive the message that the “Windows Subsystem for Linux” is missing. To install it follow these instructions
- While trying to use Docker on Windows you may receive the error “Hardware assisted virtualization and data execution protection must be enabled in the BIOS”. This requires you to access your computer BIOS to enable “hardware virtualization”. Mode of access and BIOS menu options vary with computer model.
- In some problematic cases, you may consider using an older version of Docker. See: https://docs.docker.com/desktop/release-notes/
Install R
Add Repositories
Open a local R Console and add these two additional Repositories.
options(
repos = c(
getOption("repos"),
PANDORA = "https://Pandora-IsoMemo.github.io/drat/"
INWTlab = "https://inwtlab.github.io/drat/"
)
)Install Package
Load and Install the R-Package
install.packages("ReSources")Start the app
Load and attach package
library("ReSources")Run the app
ReSources::startApplication()TraceR
GitHub Repository Documentation Start App Start App (Beta)
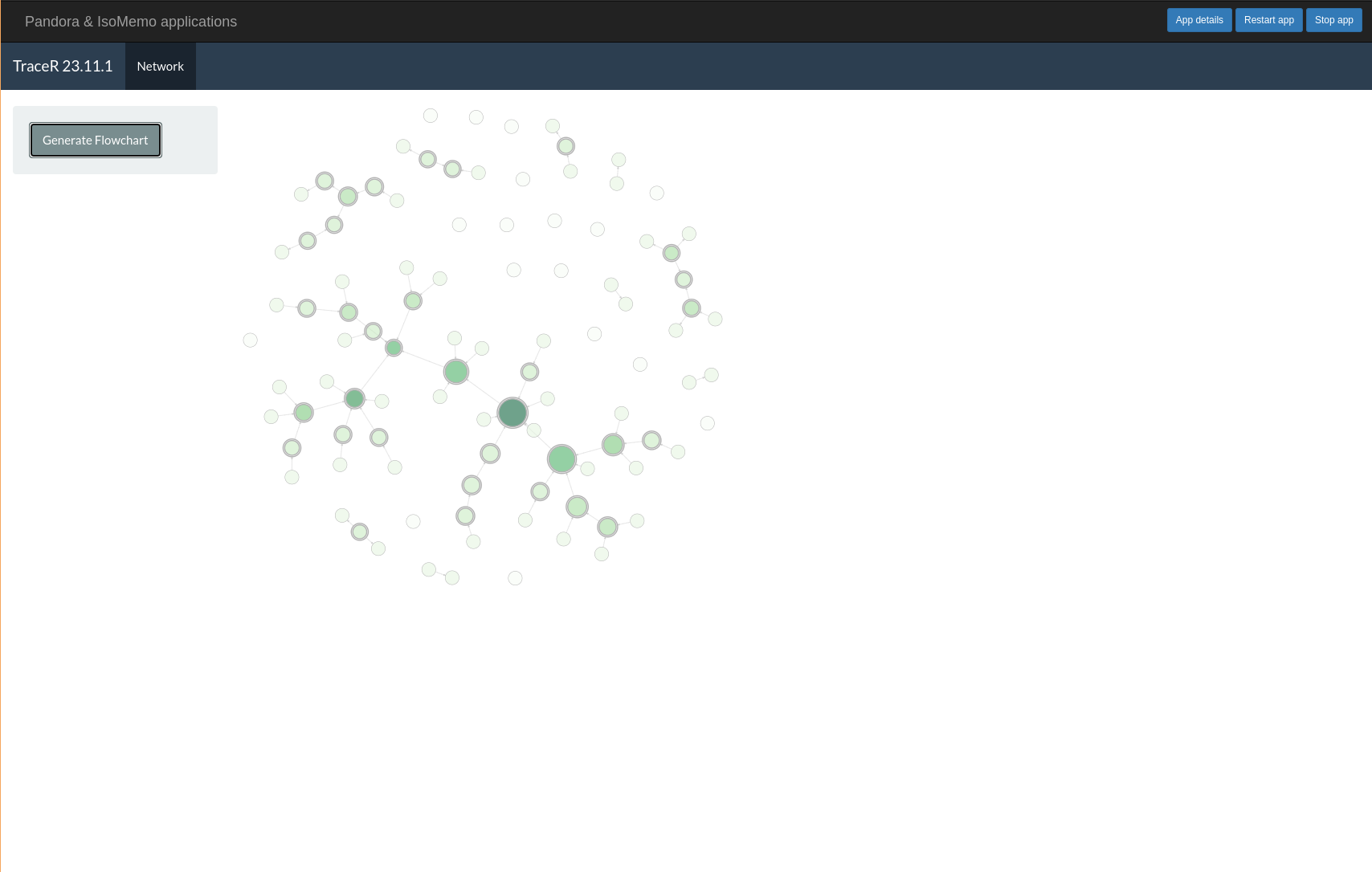
TraceR helps to generate a visual representation of a relational map.
1. Install the software Docker
Download installation files from one of the links below and follow installation instructions:
After Docker is installed you can pull & run the app manually or use the all-in-one-app.sh-Script.
2. Download and install docker image of the app
This image contains all elements necessary for you to run the app from a web browser. Run this code in a local terminal
Open a terminal (command line):
- Windows command line:
- Open the Start menu or press the
Windows key+R; - Type cmd or cmd.exe in the Run command box;
- Press Enter.
- Open the Start menu or press the
- MacOS: open the Terminal app.
- Linux: most Linux systems use the same default keyboard shortcut to start the command line:
Ctrl-Alt-TorSuper-T
Copy paste the text below into the terminal and press Enter:
docker pull ghcr.io/pandora-isomemo/tracer:main3. Run the application in Docker
Steps 1 and 2 install the app. To run the app at any time after the installation open a terminal (as described in point 2) copy paste the text below into the terminal and press Enter. Wait for a notification that the app is in “listening” mode.
docker run -p 3838:3838 ghcr.io/pandora-isomemo/tracer:mainIf the app is shutdown on Docker or if the terminal is closed the app will no longer work in your web browser (see point 4).
4. Display the app in a web browser
Once the app is running in Docker you need to display it in a web browser. For this, copy-paste the address below into your web browser’s address input and press Enter.
http://127.0.0.1:3838/Troubleshooting
- You may not be able to install Docker due a lack of administrative rights. If so, contact your IT department to grant installation rights.
- While trying to use Docker on Windows you may receive the message that the “Windows Subsystem for Linux” is missing. To install it follow these instructions
- While trying to use Docker on Windows you may receive the error “Hardware assisted virtualization and data execution protection must be enabled in the BIOS”. This requires you to access your computer BIOS to enable “hardware virtualization”. Mode of access and BIOS menu options vary with computer model.
- In some problematic cases, you may consider using an older version of Docker. See: https://docs.docker.com/desktop/release-notes/
Install R
Add Repositories
Open a local R Console and add these two additional Repositories.
options(
repos = c(
getOption("repos"),
PANDORA = "https://Pandora-IsoMemo.github.io/drat/"
INWTlab = "https://inwtlab.github.io/drat/"
)
)Install Package
Load and Install the R-Package
install.packages("TraceR")Start the app
Load and attach package
library("TraceR")Run the app
TraceR::startApplication()